This guide will give you a brief overview of how to use JBrowse for exploring the sunflower genome annotations. For reference, there is a more detailed guide for how to use JBrowse on ARAPORT (Arabidopsis Information Portal). This page serves to explain some of the features of the sunflower browser that might not be found on other sites.
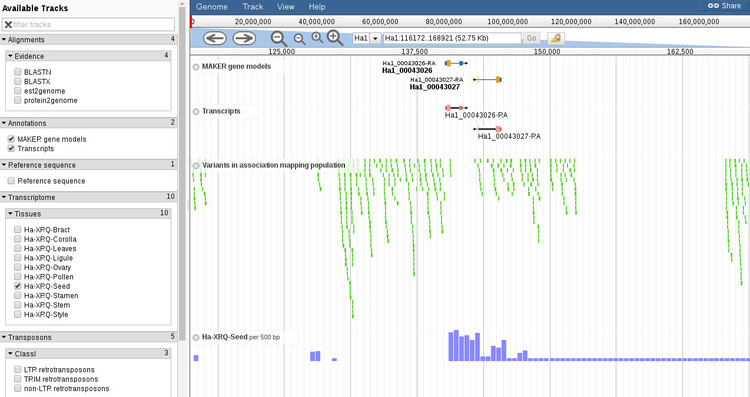
By default, the browser will open with the features shown above. You can add or remove features with the menu to the left of the browser window and navigation of the chromosome is handled with the controls at the top of the browser. If we click on a gene or transcript we can find annotation details about this feature and also export various subfeatures as FASTA. Shown below is the view when you hover over gene to open up a new window.
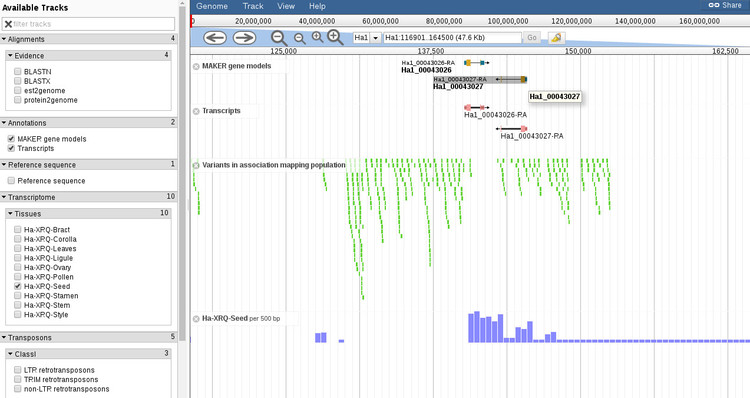
If we click on this gene it will open up a new window, as shown below.
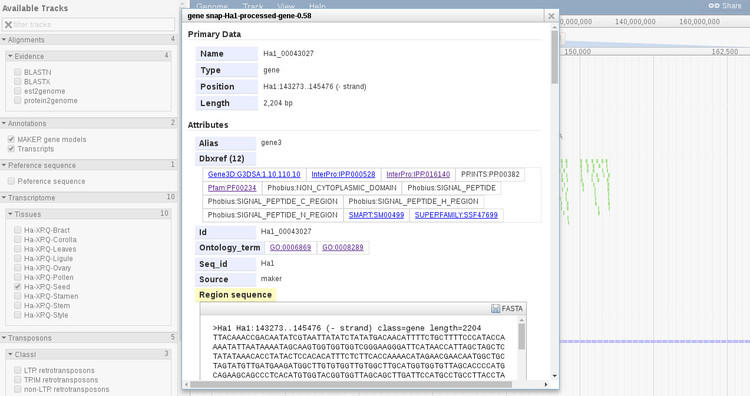
In this information panel we can see basic information such as the gene name and length, and all of the associated GO terms and other databases cross-references (under the section Dbxref). All of these links will open in a new window. If you scroll down there are is more information about the subfeatures of the gene or transcript and options to export each as FASTA.
The same options are also available for other annotated features such as transposons. You can get the name of the feature and other associated information such as the Superfamily or Family in the case of transposons, and also export subfeatures.
