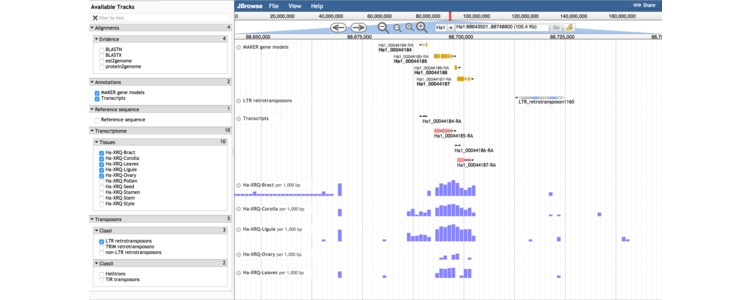
Shown above is a screen capture from the genome browser displaying expression levels of genes and transposons in a small window on chromosome 1. The transposon annotations in the genome browser and, available for download on this site, were generated with a tool called Tephra which is discussed on this page.
The primary challenge with the assembly and annotation of plant genomes is dealing with repetitive elements such as transposons. The sunflower genome has been particularly challenging due to nature of transposons. That is, most LTR retrotransposons in sunflower are very young, having accumulated since the origin of the species. While much is know about LTR retrotransposons, less attention has been given to other transposon types, in part due to a lack of published tools and methods.
The command line toolkit I have developed, called Tephra, aims to characterize all transposon types in the genome, as well as the structure and diversity of these elements in order to better infer their evolutionary history.
The usage of this tool follows Unix convention where you have a single command with subcommands that perform different functions (similar to samtools). To give an idea of the different utilities provided there is overview of the commands and their description below.
| Subcommand | Description |
|---|---|
| all | Run all subcommands and generate annotations for all transposon types |
| classifyltrs | Classify LTR retrotransposons into superfamilies and families |
| classifytirs | Classify TIR transposons into superfamilies |
| findhelitrons | Find Helitons in a genome assembly |
| findltrs | Find LTR retrotransposons in a genome assembly |
| findnonltrs | Find non-LTR retrotransposons in a genome assembly |
| findtirs | Find TIR transposons in a genome assembly |
| findtrims | Find TRIM retrotransposons in a genome assembly |
| illrecomb | Characterize the distribution of illegitimate recombination in a genome |
| ltrage | Calculate the age distribution of LTR retrotransposons |
| maskref | Mask a reference genome with transposons |
| reannotate | Transfer annotations from a reference set of repeats to Tephra annotations |
| sololtr | Find solo-LTRs in a genome assembly |
| tirage | Calculate the age distribution of TIR transposons |
This tool is open source and is available on github, where you can find information about installation and usage.
