This guide will give you a brief overview of how to use the web-based BLAST tool to find sequences of interest in the sunflower genome.
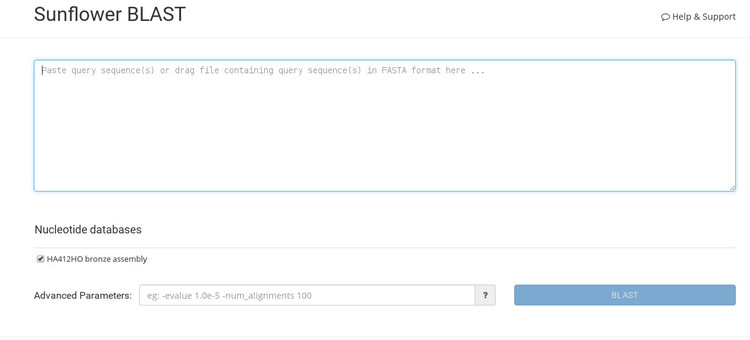
The interface for the BLAST tool is pretty straightforward (as shown in the screenshot below). Following some clues in each field, you can see there is place to paste your sequence, an optional box below this field allows you to customise the BLAST search, and finally there is a "BLAST" button to launch the job.
After pasting in our sequence and starting the job, we see the results page shown below.
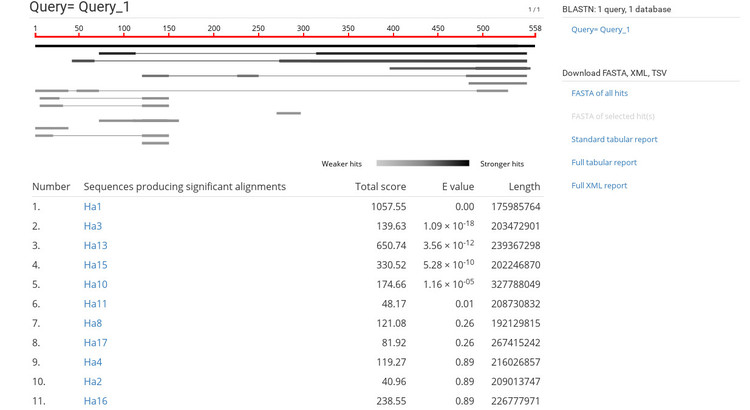
The BLAST results are very similar to NCBI's BLAST results so this should be familiar. If you click on a sequence ID in the list (under the heading "Sequences producing significant alignments") it will take you to the alignment view shown below.
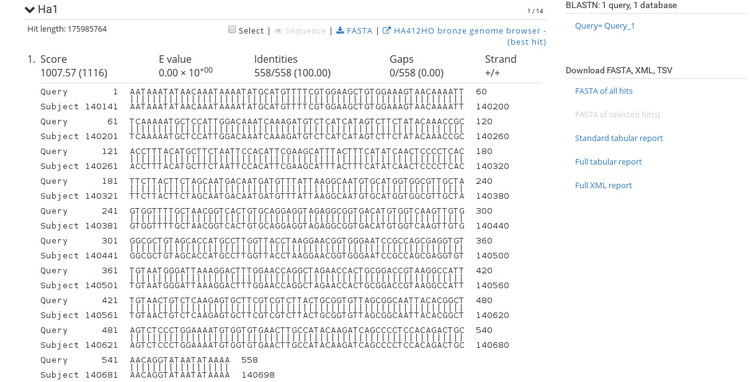
The alignment view shows basic statistics about the identity of the sequences. To the right are options for exporting the BLAST report in various formats so that you can include these results into an existing workflow. At the top of each alignment view is the option to export the hit as FASTA, or open the aligned region in the genome browser. If we click the link to open the browser, we get an image like the one shown below.
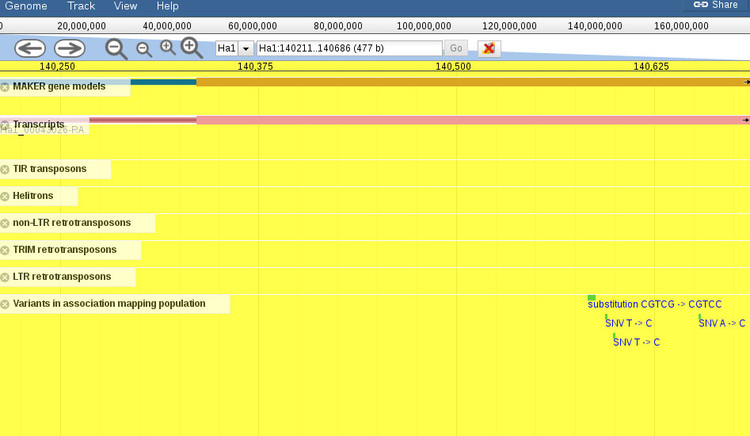
By default, the browser will open to the exact coordinates of the BLAST hit. The region will be highlighted yellow so you can zoom out (with the controls at the top of the screen) to get more context about the region, as shown below.
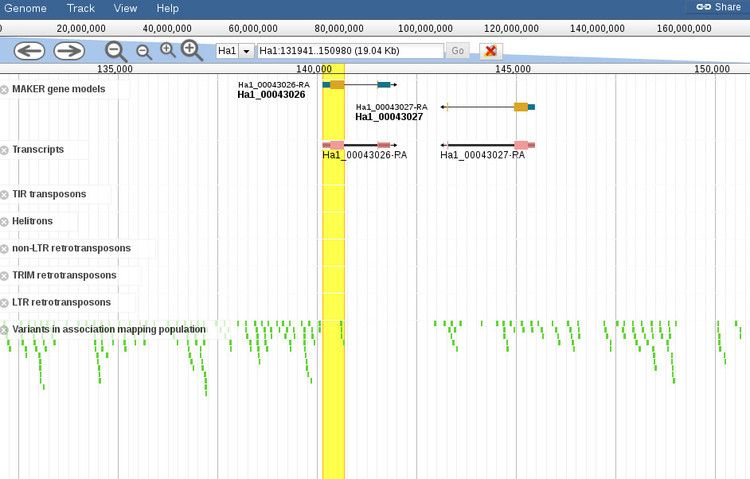
When the browser opens there will be a default set of features displayed for the region. You can add or remove any features you want by using the menu to the left of the screen (not shown in this screenshot). It should be noted that zooming out too far will cause the variants track, which is displayed by default, to not render all of the features since it is not possible at certain levels. A message will shown to indicate that variants cannot be displayed if this is the case.
