This guide will give you a brief overview of how to use the sunflower genetic map viewer cMap. There is also a more complete tutorial online that provides more details on how to use cMap.
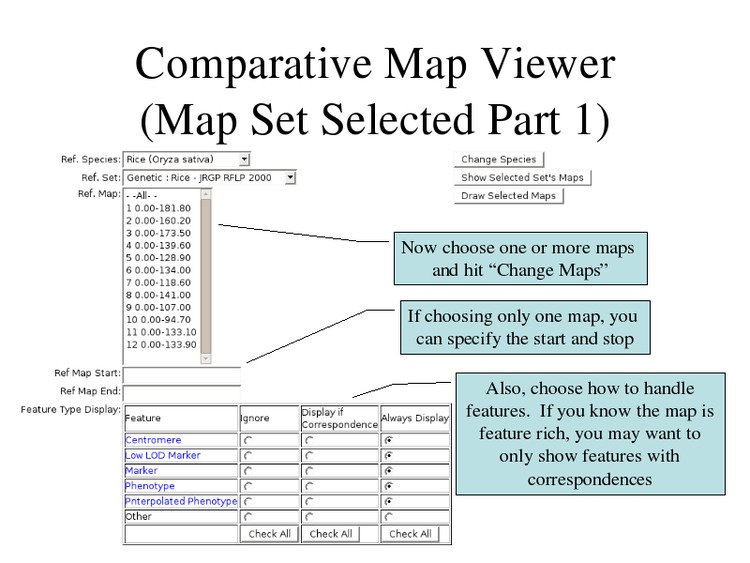
Select one or many chromosomes to draw, which features to display, and click "Draw Selected Maps".
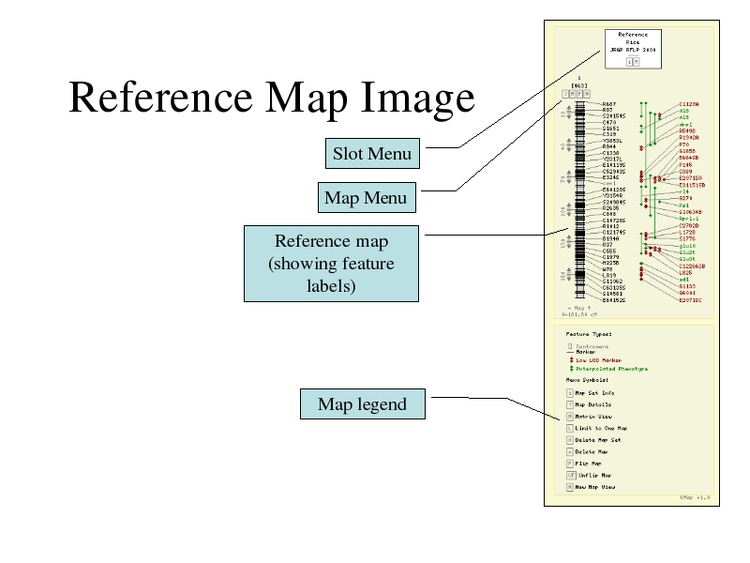
The selected maps will be displayed.
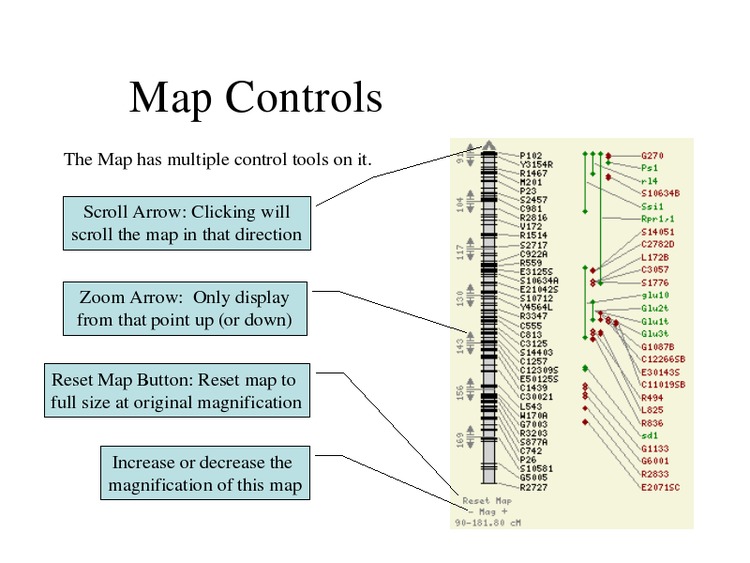
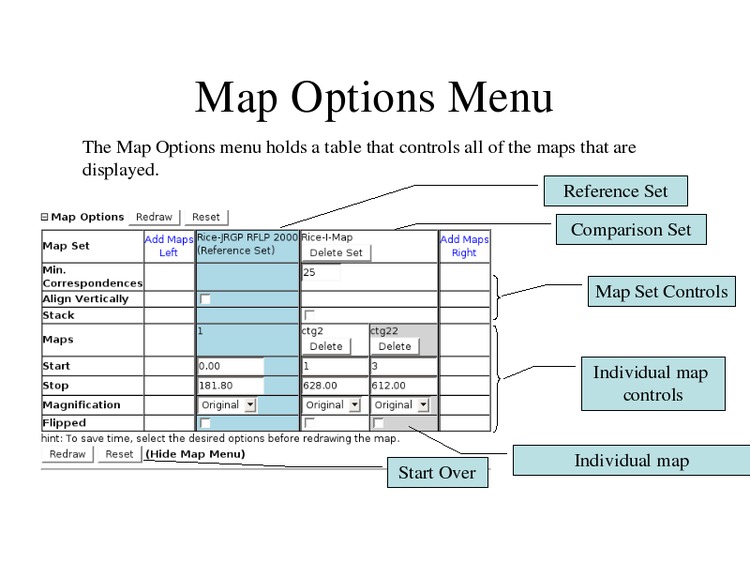
Change the map display options.
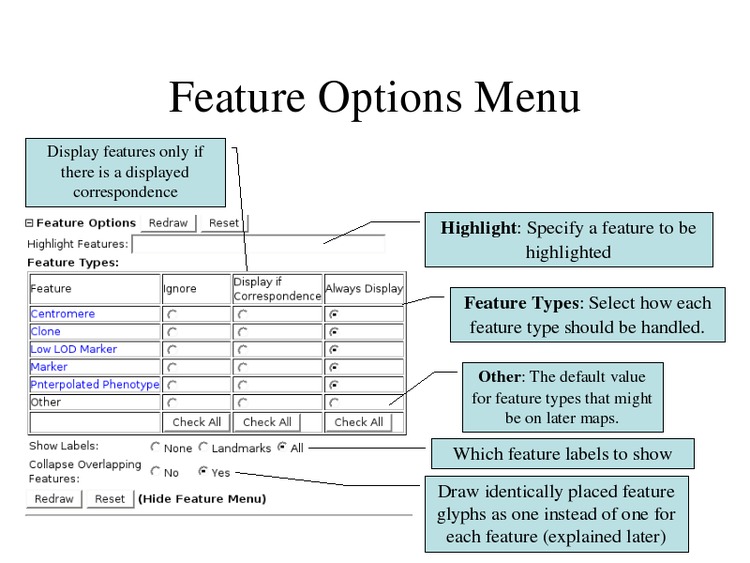
Select "All" for the Show Labels option to display all feature names.
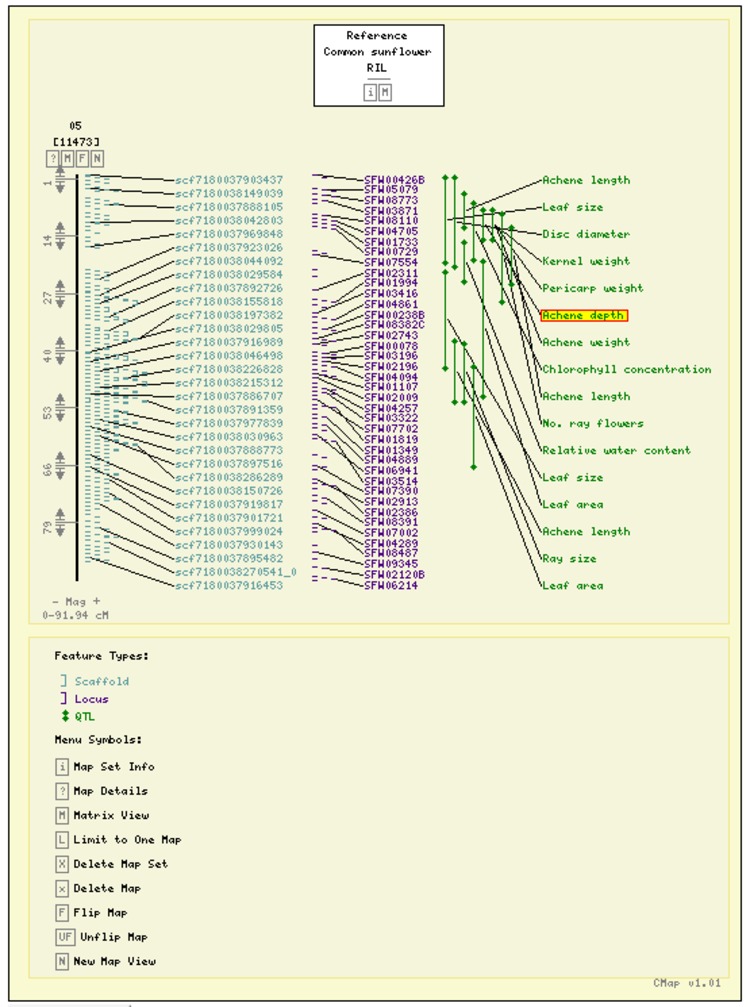
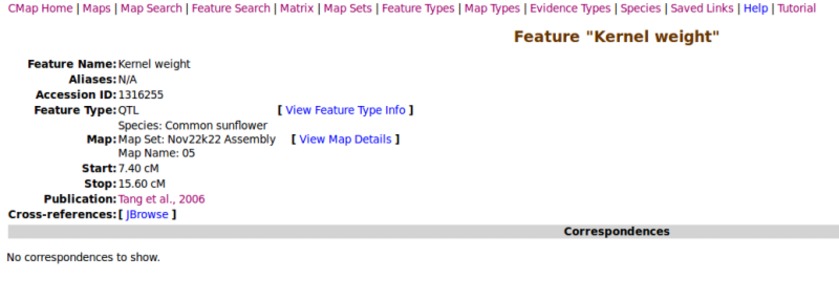
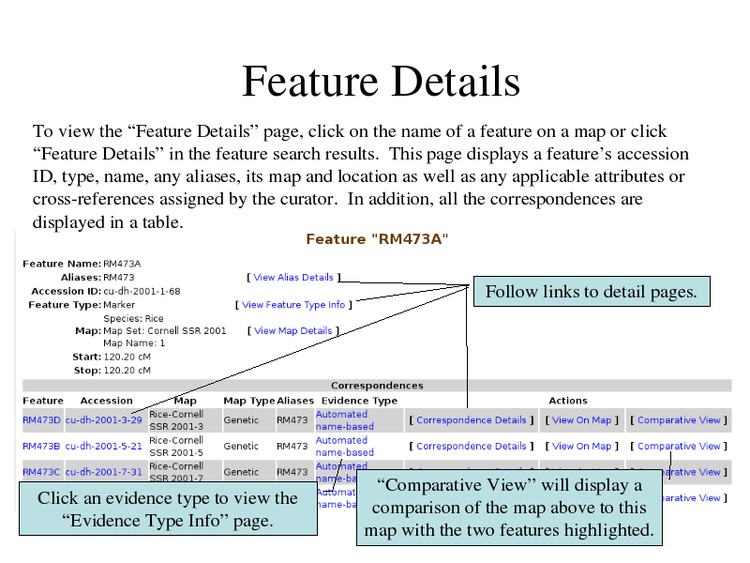
Click on a feature to view its details.
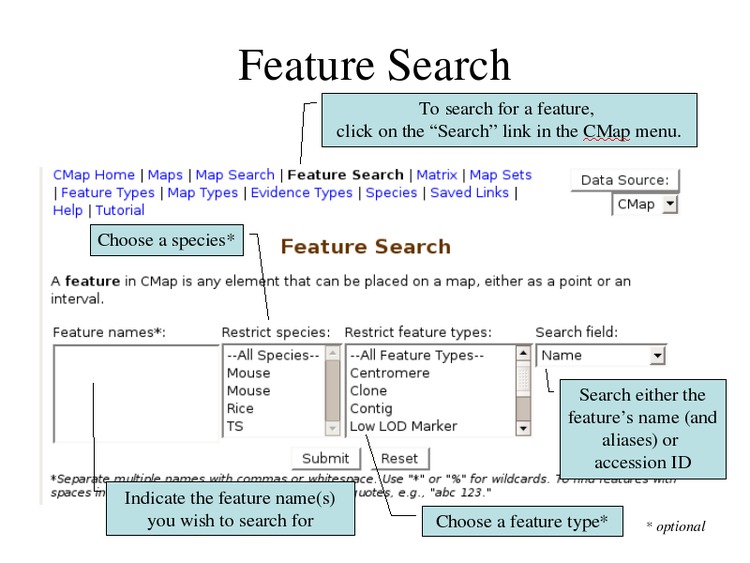
Search for individual features by name using wildcards.
Each feature is cross-linked back to its physical genome location in JBrowse.