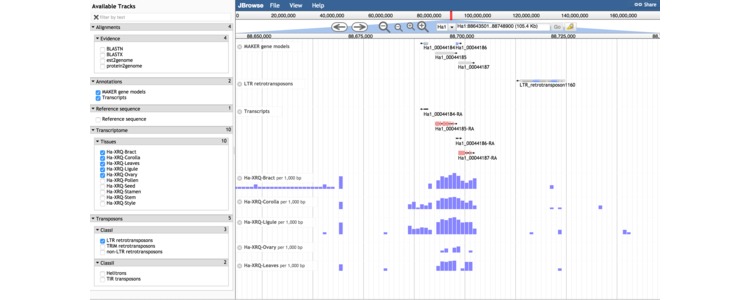
Shown above is a screen capture from the genome browser displaying variants in the association mapping population and gene expression of a gene involved in lipid storage in seeds.
I've added quite a few features to the genome browser in the past few months so I wanted to briefly go through some of these changes. First, there are some cosmetic changes that I think add to the user experience. We were previously displaying genes as a 'span' which did not show any features, as shown below.
The way genes are currently being displayed is with HTML Canvas Features in JBrowse, which highlights all of the gene features. The annotations are the same, but the way the genes are displayed in the top image adds more context and is visually nicer.
Second, the annotation cross-references (e.g., Pfam, InterPro, GO) for the transcripts and genes are now accessible in the browser. We had a subset of the annotations linking out to external databases previously, but now it should be possible to get information about all of the GO terms, Pfam domains, structural information, etc. by clicking on the gene or transcript. This was accomplished with a little JavaScript, which I've put online here and here and included minified versions for JBrowse.
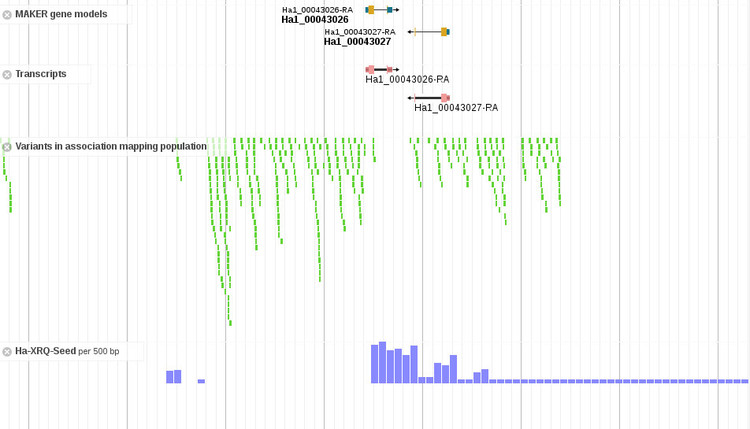
Third, there is now a "Variants" track in JBrowse for viewing variants across the association mapping population of 288 breeding lines and wild accessions. The details of this usage will be the subject of another post.
Last, to make sure all of these features work well into the future, I've updated JBrowse to latest version (1.12.1).
